
Spatial Genomics combines imaging with advanced molecular barcoding
Sequential fluorescence in situ hybridization (seqFISH) is an advanced biomolecular analysis method that merges imaging with molecular barcoding. The GenePS spatial platform uses seqFISH and makes it easy to image and decode complex molecular identities and locations directly within single cells and intact tissue microenvironments.
seqFISH enables highly multiplexed single-cell analyses of RNA, DNA, and protein beyond the capabilities of other types of spatial analyses. This technology offers the capability to determine cell types, states, and relationships by detecting and identifying dozens to tens of thousands of biomolecules while preserving intact single cells and spatial tissue organization.
How seqFISH works
With seqFISH, transcripts are identified by a fluorescent barcode that is built up over multiple images. This series of fluorescent signals uniquely identifies the gene and allows hundreds to thousands of genes to be distinguished efficiently. seqFISH has also been adapted to study the nuclear architecture of DNA and can be combined with sequential immunofluorescence for protein detection.
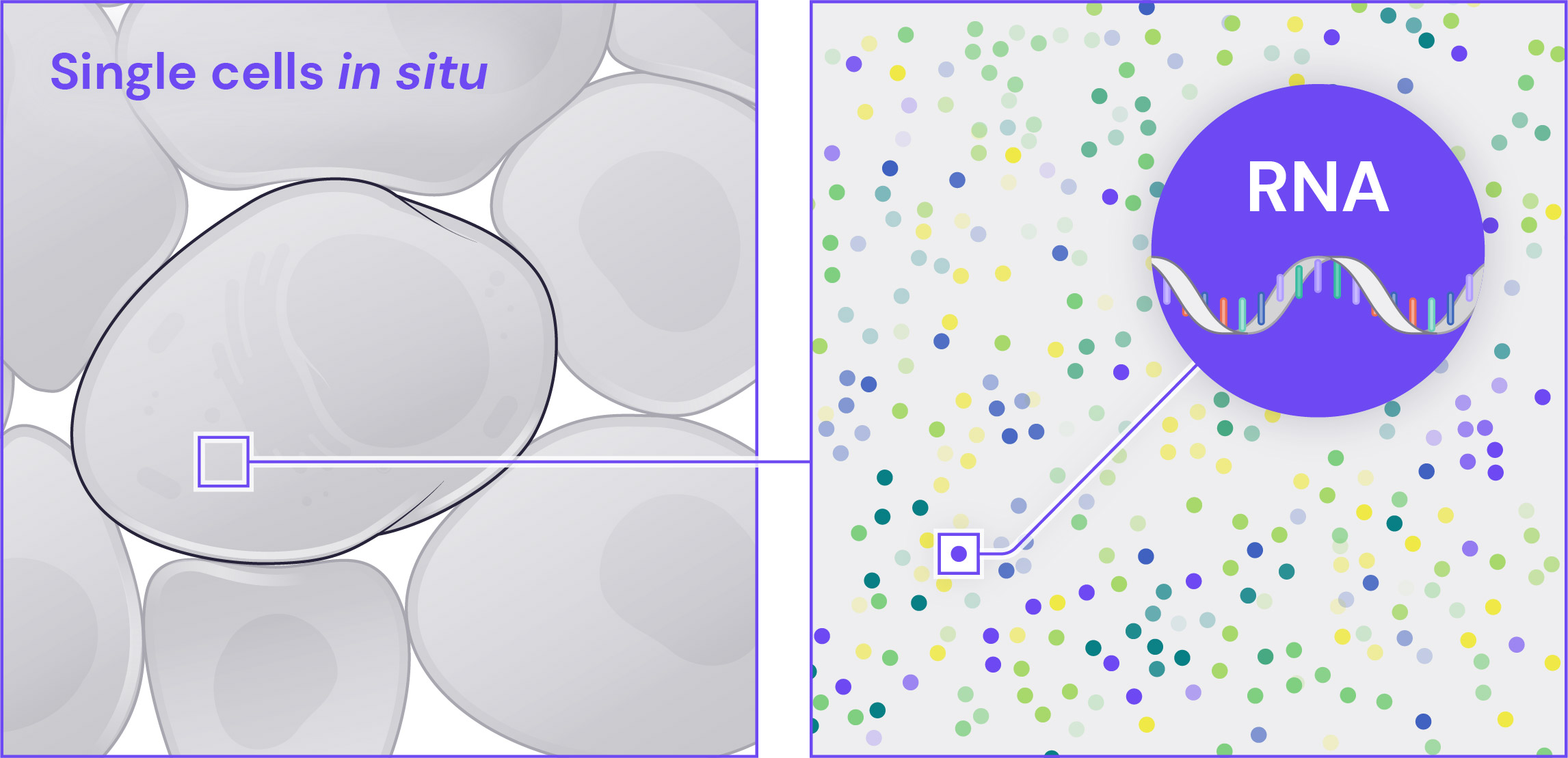
Single-Molecule RNA Specificity
Each mRNA molecule is targeted by multiple independent and highly curated probes, thereby improving signal to noise, detection, and sensitivity. The probes that detect a single mRNA molecule all fluoresce in the same channel and the same cycle of the barcoding procedure, providing robust single-molecule detection and identification.
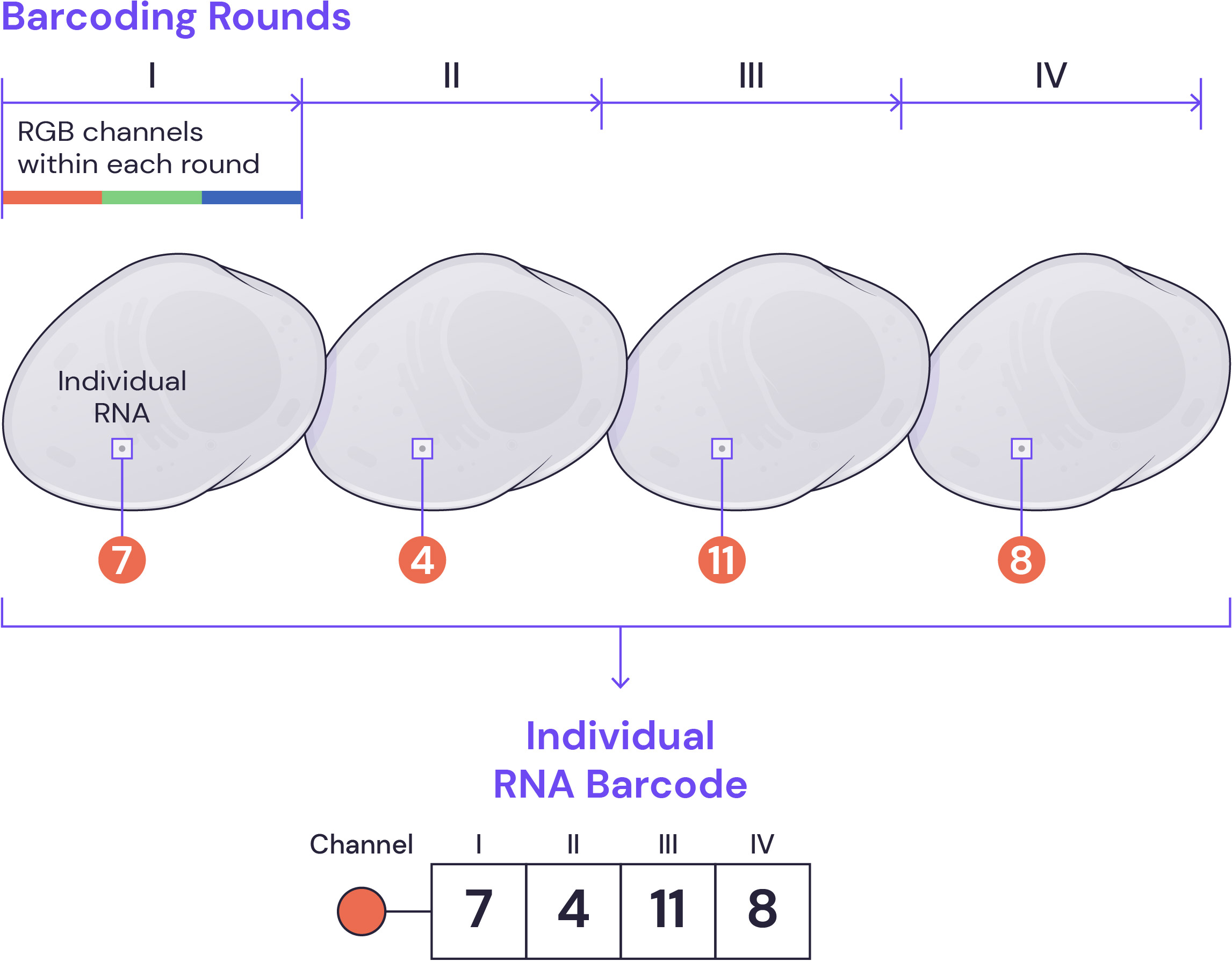
Barcoding procedure
During each round of barcoding, fluorescently tagged secondary oligonucleotide probes are hybridized to specific decoding sequences associated with each gene, imaged, and stripped. The process is repeated to build up a complete barcode, which ultimately provides a high-fidelity transcriptome map in tissues and cells with single-molecule resolution.
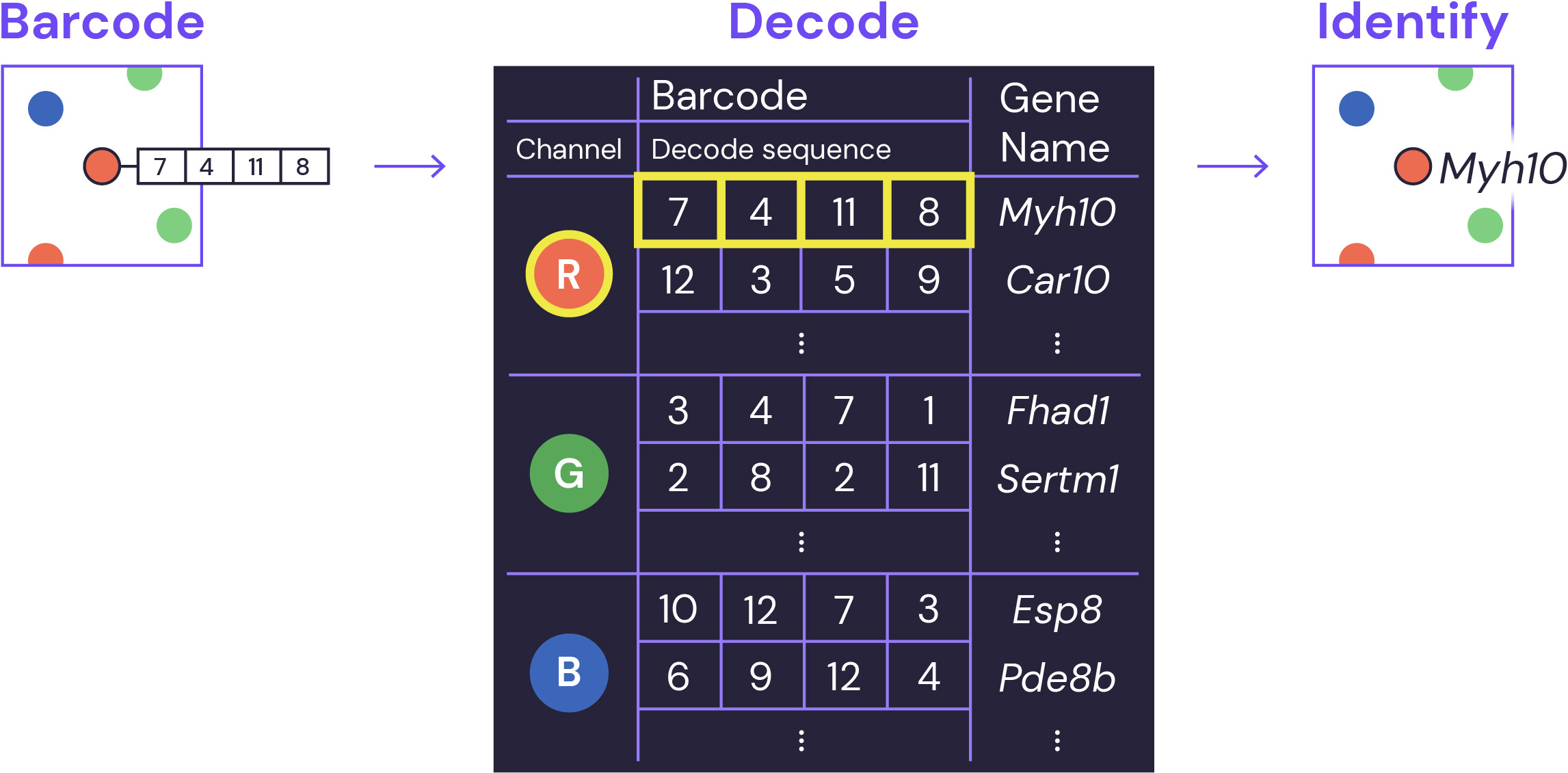
Decoding and identification
At the end of multiple barcoding rounds, the series of observed fluorescent dots provides the key for precise gene identification using an error correcting encoding scheme. All of this is made easy with our analysis software.
Why use seqFISH
Cells are molecularly crowded spaces, densely packed with genetic and biomolecular information. To reduce optical crowding, the GenePS sequentially probes a selection of biomolecules in the same cells over several rounds of imaging. With this approach, seqFISH makes possible a highly multiplexed and even full transcriptomic analysis, potentially combined with protein, DNA, or other biomolecule detection, within intact tissue sections.
All tissues and organs are composed of a complex mixture of cell types with distinct functions, environments, and stages of development. The seqFISH method yields high-resolution insights into cell types, states, relationships, and neighborhoods, within native tissue and organ environments.
Single-molecule sensitivity
seqFISH is highly sensitive because it is a direct single-molecule quantitation of nucleic acid target sequences.
Reproducibility
Our results are reproducible in corresponding sections from different samples and in dissociated or cultured cells.
Specificity
seqFISH is highly specific, with probes designed and curated to limit off-target effects.
Single-cell resolution
seqFISH enables analysis of single cells within intact tissues or cultures, without cell loss or changes in cell state due to dissociation.
Multi-omics
seqFISH can assay up to a whole transcriptome, as well as proteins and genomic loci.
Flexibility
Optimized molecular barcoding and imaging technology provides highly multiplexed analysis of hundreds or thousands of genes, while avoiding optical crowding.
